How to Maximize Retriever Performance on a More Natural Dataset
Practical considerations for improving information retrieval in an IR QA system
- Prerequisites
- Shortcomings of SQuAD2.0
- A More Realistic Alternative: Natural Questions
- Query Expansion Techniques
- Passage Ranking
- Final Thoughts
If you’ve been following along on our question answering journey thus far, you now understand the basic building blocks that form the pipeline of a modern Information Retrieval-based (IR) Question Answering system, and how that system can be evaluated on the SQuAD2.0 dataset. But, as it turns out, implementing question answering for real-world use cases is a bit more nuanced than evaluating system performance against a toy dataset. In this post, we’ll explore several challenges faced by the Retriever when applying IR-QA to a more realistic dataset, as well as a few practical approaches for overcoming them.
Prerequisites
- a basic understanding of IR-QA systems (see our previous posts)
- a basic understanding of modern NLP techniques
While SQuAD has been a popular benchmark for the task of machine comprehension, there are several perceived flaws in how the dataset was constructed that render it an unfair comparison to how humans naturally seek answers to questions. Specifically, SQuAD was created through artificial crowdsourcing where annotators were presented with a Wikipedia paragraph and asked to write questions that can be answered from it. By first reading a body of text and then generating questions, the annotators had already leaked information into the questions they crafted.
The methodology used here is not ideal, because a.) many questions lack context in absence of the provided paragraph and b.) there is a high lexical overlap between passages and questions - which artificially inflates the efficacy of exact match search tools (like Elasticsearch). Consider the following example:
Question: Other than the Automobile Club of Southern California, what other AAA Auto Club chose to simplify the divide?
Answer: California State Automobile Association
This is a fundamentally different scenario from that in the real world; human curiosity often leads us to blindly seek answers from an unknown domain. The SQuAD dataset consists of questions constructed from a known domain; essentially the process is (albeit imperfectly) rigged.
In response to the criticism of SQuAD’s shortcomings, and to spur the progress of open-domain QA systems, Google released the Natural Questions (NQ) dataset in 2019. NQ consists of real, anonymized questions issued to the Google search engine, and provides an entire Wikipedia article as context that may or may not contain the answer to a given question. The inclusion of open-ended, human-written questions and the need to reason over full pages of content make building a QA system over the NQ dataset a much more realistic and challenging task than datasets before it. Here are a few example questions from NQ:
- where does the energy in a nuclear explosion come from?
- how many episodes in season 2 breaking bad?
- meaning of cats in the cradle song?
Notice how some of the NQ questions are underspecified, raw, and syntactically erroneous. Do these seem oddly familiar to how you interact with search engines every day? Let’s explore a couple of techniques that might help overcome the challenges presented by this dataset and improve the Elasticsearch Retriever we built for our previous blog post.
As we learned previously, the inverted index data structure underlying Elasticsearch doesn’t preserve word order in a query by default. Consider the following question:
Question: "Who is the bad guy in The Hunger Games?"
In this example, we as humans can intuit that the combination of words “The Hunger Games” has a very specific meaning in contrast to the three tokens ("the", "hunger", "games") independently. We want to enable Elasticsearch to identify content specific to "The Hunger Games” trilogy, and not become entangled with general content about hunger and games. To accomplish this, we can apply named entity recognition (NER) - an information extraction technique that automatically identifies named entities (e.g., people, places, organizations, locations, etc.) in free text.
Implementing entity enrichment requires extended use of Elasticsearch’s rich query language and a pre-trained NER model (we chose one readily available through the spaCy NLP library). The process is as follows:
- Apply NER to process a question and extract out any named entities.
- Create a phrase subquery for each entity to preserve the order of tokens for that phrase in match criteria.
- Create a standard match subquery for the original question itself.
- Combine all subqueries into a boolean compound query that scores candidate documents according to overlap criteria from both question and phrase queries.
To simplify query expansion testing, we built a QueryExpander class (hidden below) that automates several query expansion methods. Let's take a look how our query is transformed through entity enrichment:
# collapse-hide
# install dependencies
!pip install elasticsearch_dsl
!pip install transformers==2.11.0
# import packages
import re
import os
import json
import time
import spacy
from elasticsearch_dsl import Q, Search
import gensim.downloader as api
from transformers import pipeline, AutoTokenizer
import warnings
warnings.simplefilter(action='ignore', category=FutureWarning)
# initialize models
nlp = spacy.load("en_core_web_sm")
word_vectors = api.load("glove-wiki-gigaword-50")
unmasker = pipeline('fill-mask', model="bert-base-uncased", tokenizer="bert-base-uncased")
tokenizer = AutoTokenizer.from_pretrained("bert-base-uncased", use_fast=True)
class QueryExpander:
'''
Query expansion utility that augments ElasticSearch queries with optional techniques
including Named Entity Recognition and Synonym Expansion
Args:
question_text
entity_args (dict) - Ex. {'spacy_model': nlp}
synonym_args (dict) - Ex. {'gensim_model': word_vectors, 'n_syns': 3} OR
{'MLM': unmasker, 'tokenizer': base_tokenizer, 'n_syns': 3, 'threshold':0.3}
'''
def __init__(self, question_text, entity_args=None, synonym_args=None):
self.question_text = question_text
self.entity_args = entity_args
self.synonym_args = synonym_args
if self.synonym_args and not self.entity_args:
raise Exception('Cannot do synonym expansion without NER! Expanding synonyms\
on named entities reduces recall.')
if self.synonym_args or self.entity_args:
self.nlp = self.entity_args['spacy_model']
self.doc = self.nlp(self.question_text)
self.build_query()
def build_query(self):
# build entity subquery
if self.entity_args:
self.extract_entities()
# identify terms to expand
if self.synonym_args:
self.identify_terms_to_expand()
# build question subquery
self.construct_question_query()
# combine subqueries
sub_queries = []
sub_queries.append(self.question_sub_query)
if hasattr(self, 'entity_sub_queries'):
sub_queries.extend(self.entity_sub_queries)
query = Q('bool', should=[*sub_queries])
self.query = query
def extract_entities(self):
'''
Extracts named entities using spaCy and constructs phrase match subqueries
for each entity. Saves both entities and subqueries as attributes.
'''
entity_list = [entity.text.lower() for entity in self.doc.ents]
entity_sub_queries = []
for ent in entity_list:
eq = Q('multi_match',
query=ent,
type='phrase',
fields=['title', 'text'])
entity_sub_queries.append(eq)
self.entities = entity_list
self.entity_sub_queries = entity_sub_queries
def identify_terms_to_expand(self):
'''
Identify terms in the question that are eligible for expansion
per a set of defined rules
'''
if hasattr(self, 'entities'):
# get unique list of entity tokens
entity_terms = [ent.split(' ') for ent in self.entities]
entity_terms = [ent for sublist in entity_terms for ent in sublist]
else:
entity_terms = []
# terms to expand are not part of entity, a stopword, numeric, etc.
entity_pos = ["NOUN","VERB","ADJ","ADV"]
terms_to_expand = [idx_term for idx_term in enumerate(self.doc) if \
(idx_term[1].lower_ not in entity_terms) and (not idx_term[1].is_stop)\
and (not idx_term[1].is_digit) and (not idx_term[1].is_punct) and
(not len(idx_term[1].lower_)==1 and idx_term[1].is_alpha) and
(idx_term[1].pos_ in entity_pos)]
self.terms_to_expand = terms_to_expand
def construct_question_query(self):
'''
Builds a multi-match query from the raw question text extended with synonyms
for any eligible terms
'''
if hasattr(self, 'terms_to_expand'):
syns = []
for i, term in self.terms_to_expand:
if 'gensim_model' in self.synonym_args.keys():
syns.extend(self.gather_synonyms_static(term))
elif 'MLM' in self.synonym_args.keys():
syns.extend(self.gather_synonyms_contextual(i, term))
syns = list(set(syns))
syns = [syn for syn in syns if (syn.isalpha() and self.nlp(syn)[0].pos_ != 'PROPN')]
question = self.question_text + ' ' + ' '.join(syns)
self.expanded_question = question
self.all_syns = syns
else:
question = self.question_text
qq = Q('multi_match',
query=question,
type='most_fields',
fields=['title', 'text'])
self.question_sub_query = qq
def gather_synonyms_contextual(self, token_index, token):
'''
Takes in a token, and returns specified number of synonyms as defined by
predictions from a masked language model
'''
tokens = [token.text for token in self.doc]
tokens[token_index] = self.synonym_args['tokenizer'].mask_token
terms = self.predict_mask(text = ' '.join(tokens),
unmasker = self.synonym_args['MLM'],
tokenizer = self.synonym_args['tokenizer'],
threshold = self.synonym_args['threshold'],
top_n = self.synonym_args['n_syns'])
return terms
@staticmethod
def predict_mask(text, unmasker, tokenizer, threshold=0, top_n=2):
'''
Given a sentence with a [MASK] token in it, this function will return the most
contextually similar terms to fill in the [MASK]
'''
preds = unmasker(text)
tokens = [tokenizer.convert_ids_to_tokens(pred['token']) for pred in preds if pred['score'] > threshold]
return tokens[:top_n]
def gather_synonyms_static(self, token):
'''
Takes in a token and returns a specified number of synonyms defined by
cosine similarity of word vectors. Uses stemming to ensure none of the
returned synonyms share the same stem (ex. photo and photos can't happen)
'''
try:
syns = self.synonym_args['gensim_model'].similar_by_word(token.lower_)
lemmas = []
final_terms = []
for item in syns:
term = item[0]
lemma = self.nlp(term)[0].lemma_
if lemma in lemmas:
continue
else:
lemmas.append(lemma)
final_terms.append(term)
if len(final_terms) == self.synonym_args['n_syns']:
break
except:
final_terms = []
return final_terms
def explain_expansion(self, entities=True):
'''
Print out an explanation for the query expansion methodology
'''
print('Question:', self.question_text, '\n')
if entities:
print('Found Entities:', self.entities, '\n')
if hasattr(self, 'terms_to_expand'):
print('Synonym Expansions:')
for i, term in self.terms_to_expand:
if 'gensim_model' in self.synonym_args.keys():
print(term, '-->', self.gather_synonyms_static(term))
elif 'MLM' in self.synonym_args.keys():
print(term, '-->', self.gather_synonyms_contextual(i,term))
else:
print('Question text has no terms to expand.')
print()
print('Expanded Question:', self.expanded_question, '\n')
print('Elasticsearch Query:\n', self.query)
question = "Who is the bad guy in The Hunger Games?"
entity_args = {'spacy_model': nlp}
qe_ner = QueryExpander(question, entity_args)
qe_ner.explain_expansion(entities=True)
From the example above, we see that our NER model successfully identified "The Hunger Games" as a named entity, and the final boolean query served to Elasticsearch is comprised of two parts: one multi-match query for the raw question text, and one phrase-match query for the extracted entity.
The exact match nature of Elasticsearch is powerful and effective, but it isn't perfect. Word matching is limited in its ability to take semantically related concepts into consideration, and for the vague nature of NQ questions, will degrade the performance of our Retriever. For example, let's further consider the question from above and a supporting context passage:
Question: "Who is the bad guy in The Hunger Games?"
Context: "President Coriolanus Snow is the main antagonistic villain in The Hunger Games trilogy, and though seemingly laid-back, his demeanor hides a sadistic mind."
An exact match-based Retriever like Elasticsearch would struggle to fetch this supporting context passage because it lacks the ability to relate the concepts of "bad guy" and "villain." More sophisticated document retrieval systems (that take advantage of learned, dense representations of text) would perform better in this situation; however, these systems are non-trivial to implement and impractical to maintain. Rather, we can try to help generalize the query through synonym expansion at search time - that is, we can identify ambiguous terms in the input question and augment the query with additional synonyms for those terms.
But how do we determine what a synonym is? And how do we know which terms in the question should be expanded?
We experimented with two methods that follow the same process, but differ slightly in how synonyms are designated. The process entails:
- Identifying a set of candidate tokens. Ideally, we want to expand tokens such that additional terms serve to increase recall, while adding minimal noise and without altering the semantics of the original query. We look at every term in the question and choose to only expand nouns, verbs, adjectives, and adverbs that are not part of a named entity.
- Expanding each candidate token. Related terms can be derived in numerous ways, from traditional lexicon lookups (e.g., WordNet) to similarity measures between learned vector space representations (e.g., Word2Vec). We’ve tested the use of static word embedding similarity and masked language model predictions as proxies for generating synonymous terms (more on these methods in a bit).
- Post-processing synonyms and crafting a new query. We then filter the expanded vocabulary to remove any duplicative, non-alphanumeric, and proper noun tokens. The final set of expanded terms is used to create a new Elasticsearch query by appending the novel words to the original question text.
Let’s take a deep dive into these two expansion methods to see how they compare.
Word embeddings are real-valued, vector representations of text that capture general contextual similarities between words in a given vocabulary. They are built on the idea that similar words tend to occur together frequently and thus are learned in an unsupervised manner from vast amounts of unstructured text. Their numerical form allows for mathematical operations - a common application being vector similarity.
Since the vectors have been trained to represent the natural co-occurence of words, we extrapolate that terms corresponding to vectors with high cosine similarity are contextually synonymous. These word relationships are learned with regard to the data they are trained on, so it is critical that the training corpus for a set of embeddings is indicative of the downstream task to which the embedding vectors will be applied. For that reason, we made use of 100 length GloVe word vectors trained on Wikipedia, available through the Gensim library.
Let's take a look at an example question and how it is expanded, using static embeddings:
question = "what is Thomas Middleditch's popular tv show?"
entity_args = {'spacy_model': nlp}
synonym_args = {'gensim_model': word_vectors,
'n_syns': 2}
qe_static = QueryExpander(question, entity_args, synonym_args)
qe_static.explain_expansion(entities=True)
Here we notice that "popular" and "tv" were the only tokens from the question deemed as candidates for expansion because all others are either stopwords or proper nouns composing a named entity. The expansion terms generated from our embedding similarity technique appear to be synonyms to the candidate terms, and by expanding the question with these related terms, we help generalize the query to capture a wider range of potentially relevant content.
However, while this example demonstrates a (mostly) successful use of embedding similarity, this is not always the case. Despite the semantic value baked into word embeddings, there are several limitations to their use as a proxy for synonymous meaning. Let's look at another example:
question = "how many rose species are found in the Montreal Botanical Garden?"
qe_static = QueryExpander(question, entity_args, synonym_args)
qe_static.explain_expansion(entities=True)
Drawing your attention to the suggested expansions for the term "rose", it becomes obvious that our approach lacks the ability to disambiguate between different senses of the same word. As humans, we naturally infer that the term "rose" implies a flower rather than the past-tense action of ascending from a lower position to a higher one. This illustrates a main limitation of word embeddings - multiple meanings of the same word are conflated as a single, static representation. In linguistic terms, polysemy and homonymy are not handled effectively.
Not only have we altered the semantics of the original query by introducing the new terms for the wrong word sense, but our approach has also failed to accurately capture synonymous terms for that mistaken word sense. We see that "rose" expanded to "fell" and "climbed"...the first of which is actually an antonym. What is going on here? As mentioned earlier, word embeddings are trained at the task of modeling co-occurrence probabilities. So while terms that occur in similar contexts may sometimes be synonymous, that certainly is not always the case.
The prior approach lacks something that allows us as humans to disambiguate the meaning of the word "rose" - context. The ability to dynamically recognize alternate meaning by paying attention to the context surrounding a term allows us to distinguish homonyms. For a computer to do the same, we can make use of a masked language model (MLM) to leverage the bi-directional context surrounding a given word and imply its morphological form.
In practice, this involves constructing intermediate versions of the original question, in which each identified candidate token is masked, and an MLM predicts the top N tokens that are contextually most likely to complete the sentence. As with static embeddings, contextual query expansion relies on the assumption that the MLM has been trained (or fine-tuned) on the target document corpus (so it holds relevant, implicit information that can be exploited to identify suitable expansion terms).
In our case, we made use of a BERT-base-uncased model that has also been pre-trained on Wikipedia, conveniently available through HuggingFace's "fill-mask" pipeline API. Let's see how this approach performs on our ambiguous example from above:
question = "how many rose species are found in the Montreal Botanical Garden?"
entity_args = {'spacy_model': nlp}
synonym_args = {'MLM': unmasker,
'tokenizer': tokenizer,
'n_syns': 2,
'threshold':0}
qe_contextual = QueryExpander(question, entity_args, synonym_args)
qe_contextual.explain_expansion(entities=True)
Great! Because we are now using a supervised model to consider the full sentence and predict the missing candidate terms, we are able to capture the correct meaning of the term "rose," and also produce more reliable "synonyms" for each of the three expanded terms.
Due to the topical breadth of Natural Questions, a vast knowledge base over which the Retriever can search is necessary in order to fairly evaluate an end-to-end QA system. For that reason, we cleaned and loaded a full Wikipedia dump into an Elasticsearch index for testing. To represent a practical application of QA, full articles were indexed rather than pre-parsed paragraphs (as we saw in the previous post). The NQ development set was processed to drop all long and yes/no answer questions, yielding 7651 short and null answer examples. Additionally, answers with more than five tokens were discarded as answers with many tokens often resemble extractive snippets rather than canonical answers.
With our knowledge corpus and evaluation data ready to go, we evaluated our Retriever performance (with the same methodology used in the last post) over an increasing number of documents, while toggling entity and synonym expansion methods at search time.
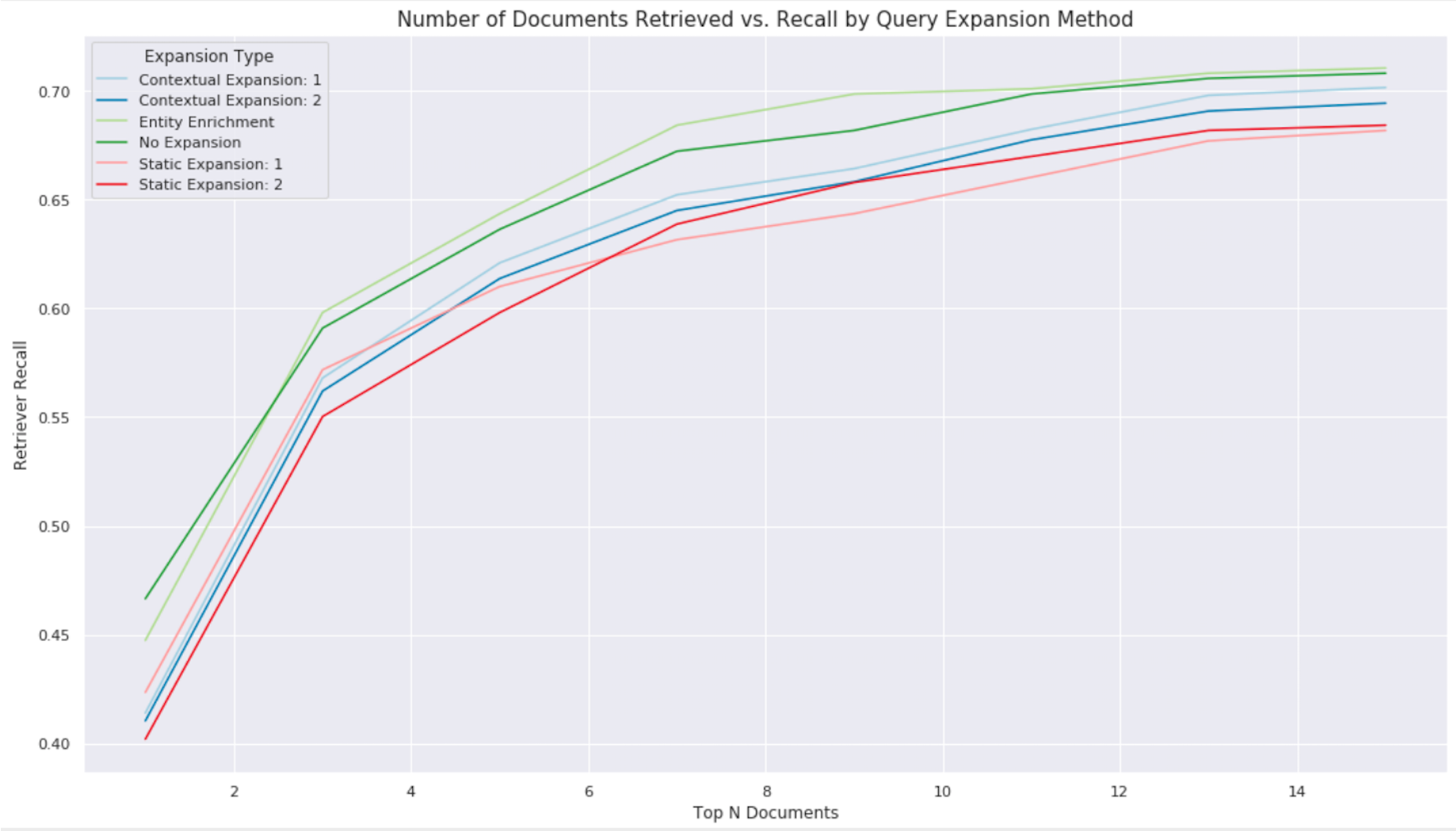
Intuitively, we see that returning more articles increases Retriever recall because more content that may contain a question’s answer exists. However, that performance gain begins to plateau as additional content becomes less relevant to the query. We observe a performance plateau occurs at ~70% recall when retrieving 14 full Wikipedia articles. Despite the differences in experimental setup, when we compare that to the ~83% recall on SQuAD in the last post (when retrieving only three paragraphs of content), it becomes evident just how much more challenging the NQ dataset actually is. We also observe that entity expansion provides a slight improvement to recall, as it increases specificity of the query definition to help target multi-word phrases in articles.
Finally, we observe a loss in performance when expanding candidate question terms with synonyms from either proposed method. While the intuition behind synonym expansion makes sense in theory, we ultimately find it very difficult to implement a "one-size-fits-all" approach for determining relevant synonyms. Even after adding a probability threshold for MLM expansion term predictions to further minimize the chance of introducing spurious terms, we are unable to consistently restrict semantic-altering words.
Qualitative analysis reveals that MLM expansion does work in many cases, but also over-generalizes in others. In the world of information retrieval, relevancy tuning is a deeply complex subject and requires customization for each use case. Therefore, it is generally not recommended to apply blanket synonym expansion techniques. Rather, expanding similar terms from a curated ontology or acronym lookup specific to your domain at search time may prove beneficial.
We know from our last post that in an IR QA system, the Reader is bounded by the performance of the Retriever, and fetching more documents increases system recall. However, simply increasing the number of documents passed to the Reader also increases the amount of irrelevant information to be processed, and degrades the overall QA system performance - in regard to both speed and accuracy. But what if we could have the best of both worlds?
Passage ranking is a technique that involves selecting a subset of re-ranked paragraphs from a collection of retrieved documents to retain the answer recall from those documents, while filtering out noisy content. By implementing a quick and efficient passage ranking technique, our QA pipeline considers more documents' worth of information, but distills content down to only the relevant pieces for the Reader to process.
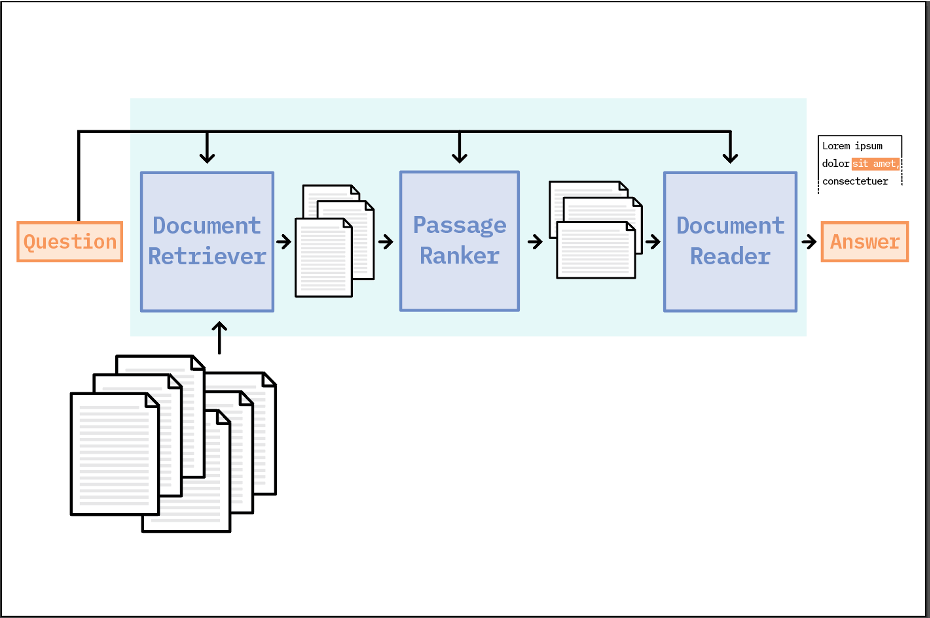
The concept of passage ranking is inspired by a field of information retrieval called Learning to Rank, which frames relevance ranking as a supervised machine learning problem. While many supervised ranking approaches have proven successful, they require learning custom similarity metrics and introduce additional complexity into a QA system - making them impractical for many use cases. In contrast, the passage ranking implementation that we consider here is a simple, unsupervised approach that demonstrates a viable way to improve IR for general question answering applications. Our passage ranking process consists of the following steps at search time:
- The query question and a set of N candidate documents from Elasticsearch are fed as input.
- All documents are split into paragraphs.
- The list of paragraphs and the input question are converted into a sparse document-term matrix (DTM) using TF-IDF vectorization. We preserve n-grams during vectorization, so the final DTM includes single terms, bi-grams, and tri-grams.
- Cosine similarity is calculated between the question vector and each paragraph vector.
- Paragraphs are sorted based on similarity score and the top M passages are passed on to the Reader for answer extraction.
To demonstrate the effect of passage ranking, we evaluated system recall across the NQ development dataset while retrieving one article’s worth of content as determined from two different methods:
- The top one document scored and returned directly from Elasticsearch
- The top 20 ranked paragraphs pooled from N candidate documents
(A fair comparison between these two methods requires consideration of an equal amount of retrieved content. We found Wikipedia articles corresponding to our NQ development set contained 20 paragraphs on average.)
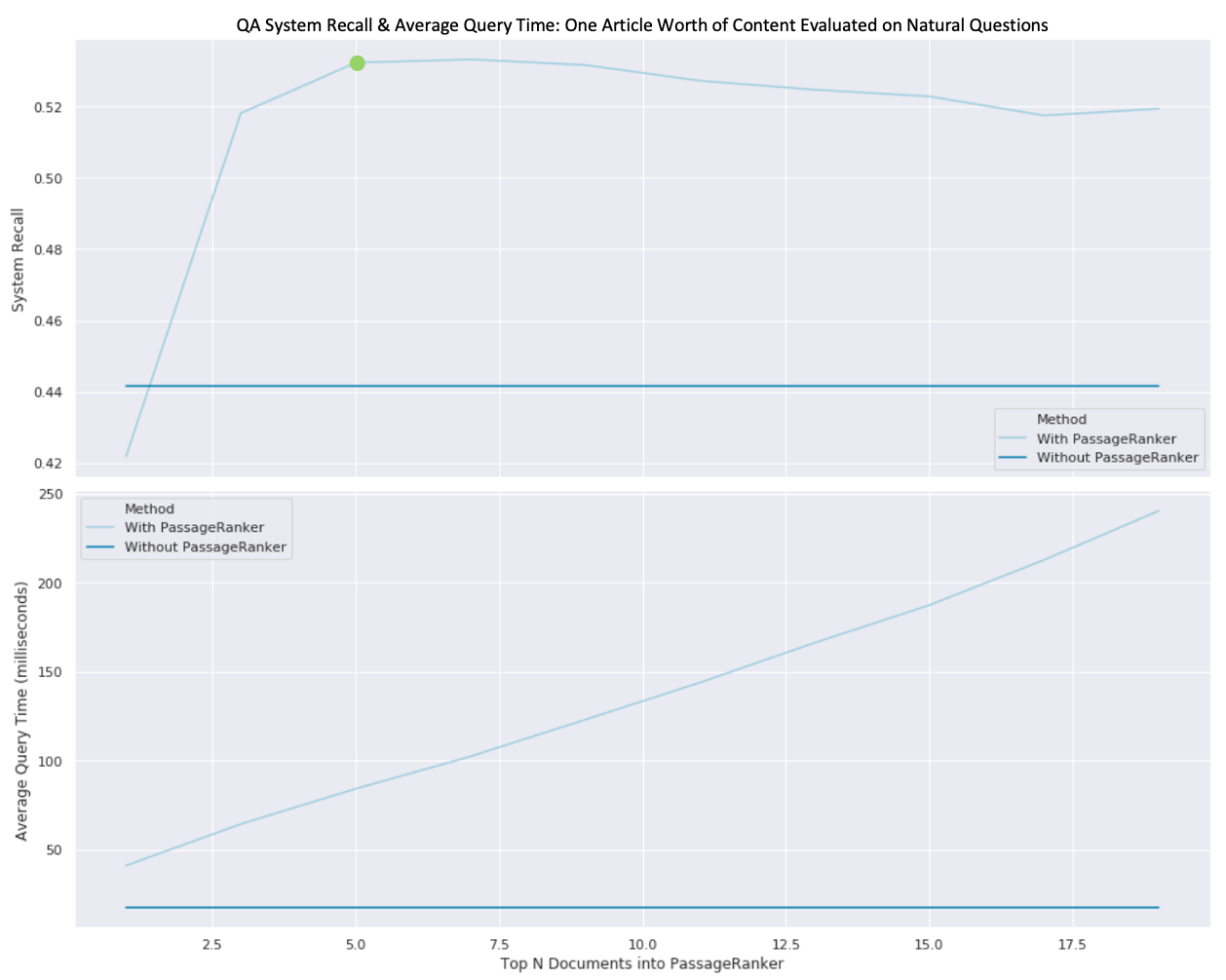
In the top half of the figure above, we see that re-ranking all paragraphs from five input documents and selecting only the top 20 (represented by the green dot) allows us to achieve a system recall of 53% in comparison to the 44% recall attained by Elasticsearch returning the top document alone (without passage ranking). These results support the notion that introducing an intermediate passage ranking step into a QA pipeline allows us to improve recall by almost ten percentage points for a fixed amount of context.
We notice that with passage ranking, recall increases as the number of candidate documents increase - up until five documents, at which point, it slowly declines. This demonstrates that after five documents' worth of content, our sparse vector ranking algorithm loses signal as additional unrelated noise is introduced to the ranking corpus.
We also evaluate the time complexity of the passage ranking process and notice that the peak recall (at five documents' worth of content) comes at a cost of four times the processing time (retrieval + passage ranking vs. just retrieving one document from Elasticsearch). While this appears considerable, it's important to frame this cost in the setting of the full QA pipeline. Passage ranking enables our already slow Reader to only process one fifth the amount of content while providing 20% more answers in the context window for the price of ~0.1 seconds.
In this post, we identified a few challenges in applying IR QA systems to a more realistic QA task, and we looked at a variety of techniques to help improve information retrieval. In the end, we found that entity expansion and passage ranking prove successful in returning more answer-containing context from which the Reader can extract answers. Additionally, we learned that while contextual synonym expansion may help Elasticsearch in some instances, it cannot be used as a blanket approach for relevancy tuning. In our final blog post, we'll explore how transfer learning can help boost Reader performance on a domain-specific dataset!